Explaining Predictions of Machine Learning Models with LIME - Münster Data Science Meetup
Slides from Münster Data Science Meetup
These are my slides from the Münster Data Science Meetup on December 12th, 2017.
knitr::include_url("https://shiring.github.io/netlify_images/lime_meetup_slides_wvsh6s.pdf")My sketchnotes were collected from these two podcasts:
- https://twimlai.com/twiml-talk-7-carlos-guestrin-explaining-predictions-machine-learning-models/
- https://dataskeptic.com/blog/episodes/2016/trusting-machine-learning-models-with-lime

Sketchnotes: TWiML Talk #7 with Carlos Guestrin – Explaining the Predictions of Machine Learning Models & Data Skeptic Podcast - Trusting Machine Learning Models with Lime
Example Code
- the following libraries were loaded:
library(tidyverse) # for tidy data analysis
library(farff) # for reading arff file
library(missForest) # for imputing missing values
library(dummies) # for creating dummy variables
library(caret) # for modeling
library(lime) # for explaining predictionsData
The Chronic Kidney Disease dataset was downloaded from UC Irvine’s Machine Learning repository: http://archive.ics.uci.edu/ml/datasets/Chronic_Kidney_Disease
data_file <- file.path("path/to/chronic_kidney_disease_full.arff")- load data with the
farffpackage
data <- readARFF(data_file)Features
- age - age
- bp - blood pressure
- sg - specific gravity
- al - albumin
- su - sugar
- rbc - red blood cells
- pc - pus cell
- pcc - pus cell clumps
- ba - bacteria
- bgr - blood glucose random
- bu - blood urea
- sc - serum creatinine
- sod - sodium
- pot - potassium
- hemo - hemoglobin
- pcv - packed cell volume
- wc - white blood cell count
- rc - red blood cell count
- htn - hypertension
- dm - diabetes mellitus
- cad - coronary artery disease
- appet - appetite
- pe - pedal edema
- ane - anemia
- class - class
Missing data
- impute missing data with Nonparametric Missing Value Imputation using Random Forest (
missForestpackage)
data_imp <- missForest(data)One-hot encoding
- create dummy variables (
caret::dummy.data.frame()) - scale and center
data_imp_final <- data_imp$ximp
data_dummy <- dummy.data.frame(dplyr::select(data_imp_final, -class), sep = "_")
data <- cbind(dplyr::select(data_imp_final, class), scale(data_dummy,
center = apply(data_dummy, 2, min),
scale = apply(data_dummy, 2, max)))Modeling
set.seed(42)
index <- createDataPartition(data$class, p = 0.9, list = FALSE)
train_data <- data[index, ]
test_data <- data[-index, ]
model_rf <- caret::train(class ~ .,
data = train_data,
method = "rf",
trControl = trainControl(method = "repeatedcv",
number = 10,
repeats = 5,
verboseIter = FALSE))model_rf# Random Forest
#
# 360 samples
# 48 predictor
# 2 classes: 'ckd', 'notckd'
#
# No pre-processing
# Resampling: Cross-Validated (10 fold, repeated 5 times)
# Summary of sample sizes: 324, 324, 324, 324, 325, 324, ...
# Resampling results across tuning parameters:
#
# mtry Accuracy Kappa
# 2 0.9922647 0.9838466
# 25 0.9917392 0.9826070
# 48 0.9872930 0.9729881
#
# Accuracy was used to select the optimal model using the largest value.
# The final value used for the model was mtry = 2.# predictions
pred <- data.frame(sample_id = 1:nrow(test_data), predict(model_rf, test_data, type = "prob"), actual = test_data$class) %>%
mutate(prediction = colnames(.)[2:3][apply(.[, 2:3], 1, which.max)], correct = ifelse(actual == prediction, "correct", "wrong"))
confusionMatrix(pred$actual, pred$prediction)# Confusion Matrix and Statistics
#
# Reference
# Prediction ckd notckd
# ckd 23 2
# notckd 0 15
#
# Accuracy : 0.95
# 95% CI : (0.8308, 0.9939)
# No Information Rate : 0.575
# P-Value [Acc > NIR] : 1.113e-07
#
# Kappa : 0.8961
# Mcnemar's Test P-Value : 0.4795
#
# Sensitivity : 1.0000
# Specificity : 0.8824
# Pos Pred Value : 0.9200
# Neg Pred Value : 1.0000
# Prevalence : 0.5750
# Detection Rate : 0.5750
# Detection Prevalence : 0.6250
# Balanced Accuracy : 0.9412
#
# 'Positive' Class : ckd
# LIME
- LIME needs data without response variable
train_x <- dplyr::select(train_data, -class)
test_x <- dplyr::select(test_data, -class)
train_y <- dplyr::select(train_data, class)
test_y <- dplyr::select(test_data, class)- build explainer
explainer <- lime(train_x, model_rf, n_bins = 5, quantile_bins = TRUE)- run
explain()function
explanation_df <- lime::explain(test_x, explainer, n_labels = 1, n_features = 8, n_permutations = 1000, feature_select = "forward_selection")- model reliability
explanation_df %>%
ggplot(aes(x = model_r2, fill = label)) +
geom_density(alpha = 0.5)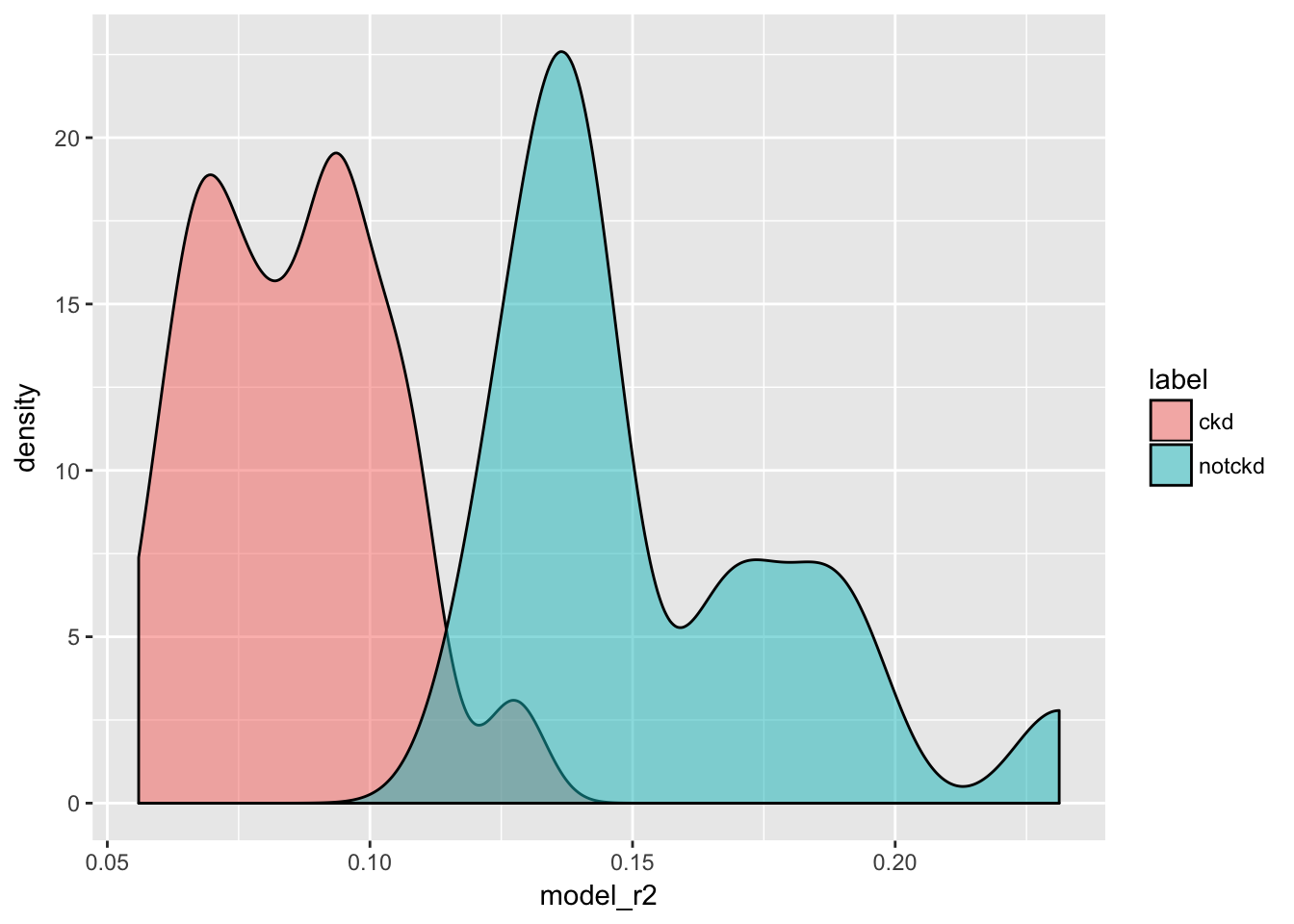
- plot explanations
plot_features(explanation_df[1:24, ], ncol = 1)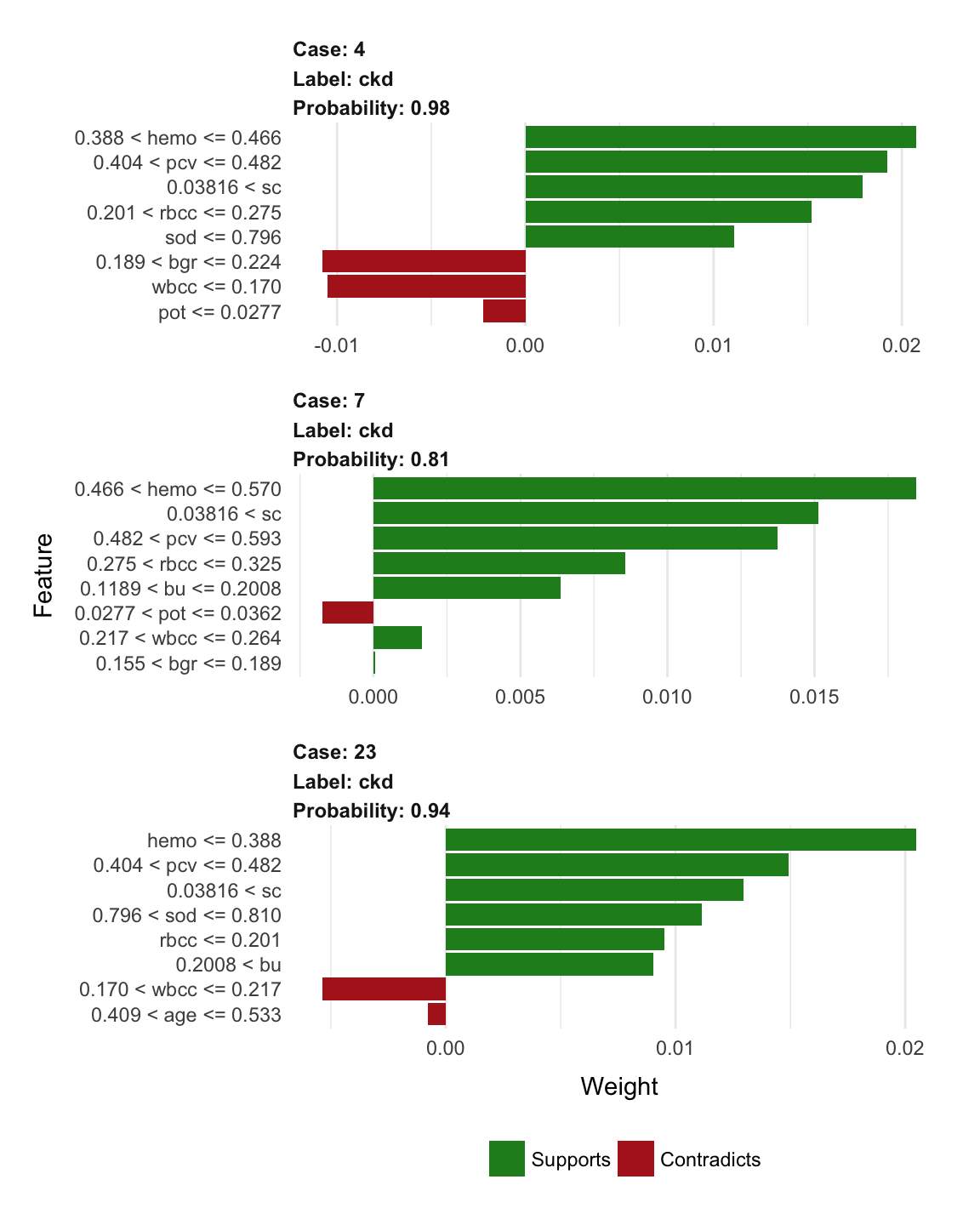
Session Info
# Session info -------------------------------------------------------------# setting value
# version R version 3.4.3 (2017-11-30)
# system x86_64, darwin15.6.0
# ui X11
# language (EN)
# collate de_DE.UTF-8
# tz Europe/Berlin
# date 2018-04-22# Packages -----------------------------------------------------------------# package * version date source
# assertthat 0.2.0 2017-04-11 CRAN (R 3.4.0)
# backports 1.1.2 2017-12-13 CRAN (R 3.4.3)
# base * 3.4.3 2017-12-07 local
# BBmisc 1.11 2017-03-10 CRAN (R 3.4.0)
# bindr 0.1 2016-11-13 CRAN (R 3.4.0)
# bindrcpp * 0.2 2017-06-17 CRAN (R 3.4.0)
# blogdown 0.5 2018-01-24 CRAN (R 3.4.3)
# bookdown 0.7 2018-02-18 CRAN (R 3.4.3)
# broom 0.4.3 2017-11-20 CRAN (R 3.4.2)
# caret * 6.0-78 2017-12-10 CRAN (R 3.4.3)
# cellranger 1.1.0 2016-07-27 CRAN (R 3.4.0)
# checkmate 1.8.5 2017-10-24 CRAN (R 3.4.2)
# class 7.3-14 2015-08-30 CRAN (R 3.4.3)
# cli 1.0.0 2017-11-05 CRAN (R 3.4.2)
# codetools 0.2-15 2016-10-05 CRAN (R 3.4.3)
# colorspace 1.3-2 2016-12-14 CRAN (R 3.4.0)
# compiler 3.4.3 2017-12-07 local
# crayon 1.3.4 2017-09-16 CRAN (R 3.4.1)
# CVST 0.2-1 2013-12-10 CRAN (R 3.4.0)
# datasets * 3.4.3 2017-12-07 local
# ddalpha 1.3.1.1 2018-02-02 CRAN (R 3.4.3)
# DEoptimR 1.0-8 2016-11-19 CRAN (R 3.4.0)
# devtools 1.13.5 2018-02-18 CRAN (R 3.4.3)
# digest 0.6.15 2018-01-28 CRAN (R 3.4.3)
# dimRed 0.1.0 2017-05-04 CRAN (R 3.4.0)
# dplyr * 0.7.4 2017-09-28 CRAN (R 3.4.2)
# DRR 0.0.3 2018-01-06 CRAN (R 3.4.3)
# dummies * 1.5.6 2012-06-14 CRAN (R 3.4.0)
# e1071 1.6-8 2017-02-02 CRAN (R 3.4.0)
# evaluate 0.10.1 2017-06-24 CRAN (R 3.4.1)
# farff * 1.0 2016-09-11 CRAN (R 3.4.0)
# forcats * 0.3.0 2018-02-19 CRAN (R 3.4.3)
# foreach * 1.4.4 2017-12-12 CRAN (R 3.4.3)
# foreign 0.8-69 2017-06-22 CRAN (R 3.4.3)
# ggplot2 * 2.2.1.9000 2018-02-28 Github (thomasp85/ggplot2@7859a29)
# glmnet 2.0-13 2017-09-22 CRAN (R 3.4.2)
# glue 1.2.0 2017-10-29 CRAN (R 3.4.2)
# gower 0.1.2 2017-02-23 CRAN (R 3.4.0)
# graphics * 3.4.3 2017-12-07 local
# grDevices * 3.4.3 2017-12-07 local
# grid 3.4.3 2017-12-07 local
# gtable 0.2.0 2016-02-26 CRAN (R 3.4.0)
# haven 1.1.1 2018-01-18 CRAN (R 3.4.3)
# highr 0.6 2016-05-09 CRAN (R 3.4.0)
# hms 0.4.1 2018-01-24 CRAN (R 3.4.3)
# htmltools 0.3.6 2017-04-28 CRAN (R 3.4.0)
# htmlwidgets 1.0 2018-01-20 CRAN (R 3.4.3)
# httpuv 1.3.6.1 2018-02-28 CRAN (R 3.4.3)
# httr 1.3.1 2017-08-20 CRAN (R 3.4.1)
# ipred 0.9-6 2017-03-01 CRAN (R 3.4.0)
# iterators * 1.0.9 2017-12-12 CRAN (R 3.4.3)
# itertools * 0.1-3 2014-03-12 CRAN (R 3.4.0)
# jsonlite 1.5 2017-06-01 CRAN (R 3.4.0)
# kernlab 0.9-25 2016-10-03 CRAN (R 3.4.0)
# knitr 1.20 2018-02-20 CRAN (R 3.4.3)
# labeling 0.3 2014-08-23 CRAN (R 3.4.0)
# lattice * 0.20-35 2017-03-25 CRAN (R 3.4.3)
# lava 1.6 2018-01-13 CRAN (R 3.4.3)
# lazyeval 0.2.1 2017-10-29 CRAN (R 3.4.2)
# lime * 0.3.1 2017-11-24 CRAN (R 3.4.3)
# lubridate 1.7.3 2018-02-27 CRAN (R 3.4.3)
# magrittr 1.5 2014-11-22 CRAN (R 3.4.0)
# MASS 7.3-49 2018-02-23 CRAN (R 3.4.3)
# Matrix 1.2-12 2017-11-20 CRAN (R 3.4.3)
# memoise 1.1.0 2017-04-21 CRAN (R 3.4.0)
# methods * 3.4.3 2017-12-07 local
# mime 0.5 2016-07-07 CRAN (R 3.4.0)
# missForest * 1.4 2013-12-31 CRAN (R 3.4.0)
# mnormt 1.5-5 2016-10-15 CRAN (R 3.4.0)
# ModelMetrics 1.1.0 2016-08-26 CRAN (R 3.4.0)
# modelr 0.1.1 2017-07-24 CRAN (R 3.4.1)
# munsell 0.4.3 2016-02-13 CRAN (R 3.4.0)
# nlme 3.1-131.1 2018-02-16 CRAN (R 3.4.3)
# nnet 7.3-12 2016-02-02 CRAN (R 3.4.3)
# parallel 3.4.3 2017-12-07 local
# pillar 1.2.1 2018-02-27 CRAN (R 3.4.3)
# pkgconfig 2.0.1 2017-03-21 CRAN (R 3.4.0)
# plyr 1.8.4 2016-06-08 CRAN (R 3.4.0)
# prodlim 1.6.1 2017-03-06 CRAN (R 3.4.0)
# psych 1.7.8 2017-09-09 CRAN (R 3.4.1)
# purrr * 0.2.4 2017-10-18 CRAN (R 3.4.2)
# R6 2.2.2 2017-06-17 CRAN (R 3.4.0)
# randomForest * 4.6-12 2015-10-07 CRAN (R 3.4.0)
# Rcpp 0.12.15 2018-01-20 CRAN (R 3.4.3)
# RcppRoll 0.2.2 2015-04-05 CRAN (R 3.4.0)
# readr * 1.1.1 2017-05-16 CRAN (R 3.4.0)
# readxl 1.0.0 2017-04-18 CRAN (R 3.4.0)
# recipes 0.1.2 2018-01-11 CRAN (R 3.4.3)
# reshape2 1.4.3 2017-12-11 CRAN (R 3.4.3)
# rlang 0.2.0.9000 2018-02-28 Github (tidyverse/rlang@9ea33dd)
# rmarkdown 1.8 2017-11-17 CRAN (R 3.4.2)
# robustbase 0.92-8 2017-11-01 CRAN (R 3.4.2)
# rpart 4.1-13 2018-02-23 CRAN (R 3.4.3)
# rprojroot 1.3-2 2018-01-03 CRAN (R 3.4.3)
# rstudioapi 0.7 2017-09-07 CRAN (R 3.4.1)
# rvest 0.3.2 2016-06-17 CRAN (R 3.4.0)
# scales 0.5.0.9000 2018-02-28 Github (hadley/scales@d767915)
# sfsmisc 1.1-1 2017-06-08 CRAN (R 3.4.0)
# shiny 1.0.5 2017-08-23 CRAN (R 3.4.1)
# shinythemes 1.1.1 2016-10-12 CRAN (R 3.4.0)
# splines 3.4.3 2017-12-07 local
# stats * 3.4.3 2017-12-07 local
# stats4 3.4.3 2017-12-07 local
# stringdist 0.9.4.6 2017-07-31 CRAN (R 3.4.1)
# stringi 1.1.6 2017-11-17 CRAN (R 3.4.2)
# stringr * 1.3.0 2018-02-19 CRAN (R 3.4.3)
# survival 2.41-3 2017-04-04 CRAN (R 3.4.3)
# tibble * 1.4.2 2018-01-22 CRAN (R 3.4.3)
# tidyr * 0.8.0 2018-01-29 CRAN (R 3.4.3)
# tidyselect 0.2.4 2018-02-26 CRAN (R 3.4.3)
# tidyverse * 1.2.1 2017-11-14 CRAN (R 3.4.2)
# timeDate 3043.102 2018-02-21 CRAN (R 3.4.3)
# tools 3.4.3 2017-12-07 local
# utils * 3.4.3 2017-12-07 local
# withr 2.1.1.9000 2018-02-28 Github (jimhester/withr@5d05571)
# xfun 0.1 2018-01-22 CRAN (R 3.4.3)
# xml2 1.2.0 2018-01-24 CRAN (R 3.4.3)
# xtable 1.8-2 2016-02-05 CRAN (R 3.4.0)
# yaml 2.1.17 2018-02-27 CRAN (R 3.4.3)